| MA1712.1_MA1712.1.ZNF454 |
MA1712.1.ZNF454,MA0811.1.TFAP2B,MA1637.1.EBF3,MA0814.2.TFAP2C,MA1604.1.EBF2, (...) |
|
-2.30 |
-2.41 |
-2.48 |
-1.90 |
2.77 |
0.69 |
2.55 |
3.33 |
-1.71 |
-1.94 |
-1.84 |
-1.73 |
3.34 |
3.34 |
3.54 |
3.55 |
1 |
0.05 |
0.06 |
0.06 |
0.06 |
-0.03 |
0.03 |
-0.01 |
-0.02 |
-0.04 |
-0.05 |
-0.05 |
-0.06 |
0.00 |
-0.01 |
-0.03 |
-0.02 |
| MA1575.1_MA1575.1.THRB |
MA1575.1.THRB,MA1597.1.ZNF528 |
|
-2.85 |
-3.03 |
-3.10 |
-2.61 |
2.61 |
0.19 |
1.60 |
2.41 |
1.27 |
1.51 |
1.33 |
1.64 |
1.82 |
1.32 |
1.62 |
1.64 |
<1 |
0.04 |
0.04 |
0.04 |
0.04 |
-0.04 |
0.03 |
-0.02 |
-0.04 |
0.02 |
0.00 |
0.00 |
-0.02 |
-0.02 |
-0.05 |
-0.06 |
-0.05 |
| MA0832.1_MA0832.1.Tcf21 |
MA0832.1.TCF21,MA0665.1.MSC,MA1621.1.RBPJL,MA0745.2.SNAI2,MA0820.1.FIGLA, (...) |
|
-3.39 |
-3.23 |
-2.81 |
-3.39 |
-0.68 |
0.20 |
0.70 |
-0.70 |
1.14 |
1.14 |
0.83 |
1.70 |
3.24 |
2.86 |
1.62 |
3.10 |
<1 |
0.05 |
0.05 |
0.06 |
0.04 |
-0.06 |
0.01 |
-0.01 |
-0.06 |
-0.02 |
-0.03 |
-0.03 |
-0.04 |
0.02 |
-0.00 |
-0.03 |
-0.01 |
| MA0484.2_MA0484.2.HNF4G |
MA0484.2.HNF4G,MA0114.4.HNF4A |
|
-4.35 |
-4.35 |
-4.35 |
-4.35 |
1.73 |
-2.71 |
-1.60 |
0.41 |
4.03 |
4.35 |
4.35 |
4.35 |
1.05 |
1.37 |
1.29 |
0.51 |
2 |
-0.05 |
-0.05 |
-0.05 |
-0.06 |
-0.03 |
-0.01 |
-0.03 |
-0.04 |
0.11 |
0.12 |
0.11 |
0.10 |
-0.01 |
-0.03 |
-0.03 |
-0.03 |
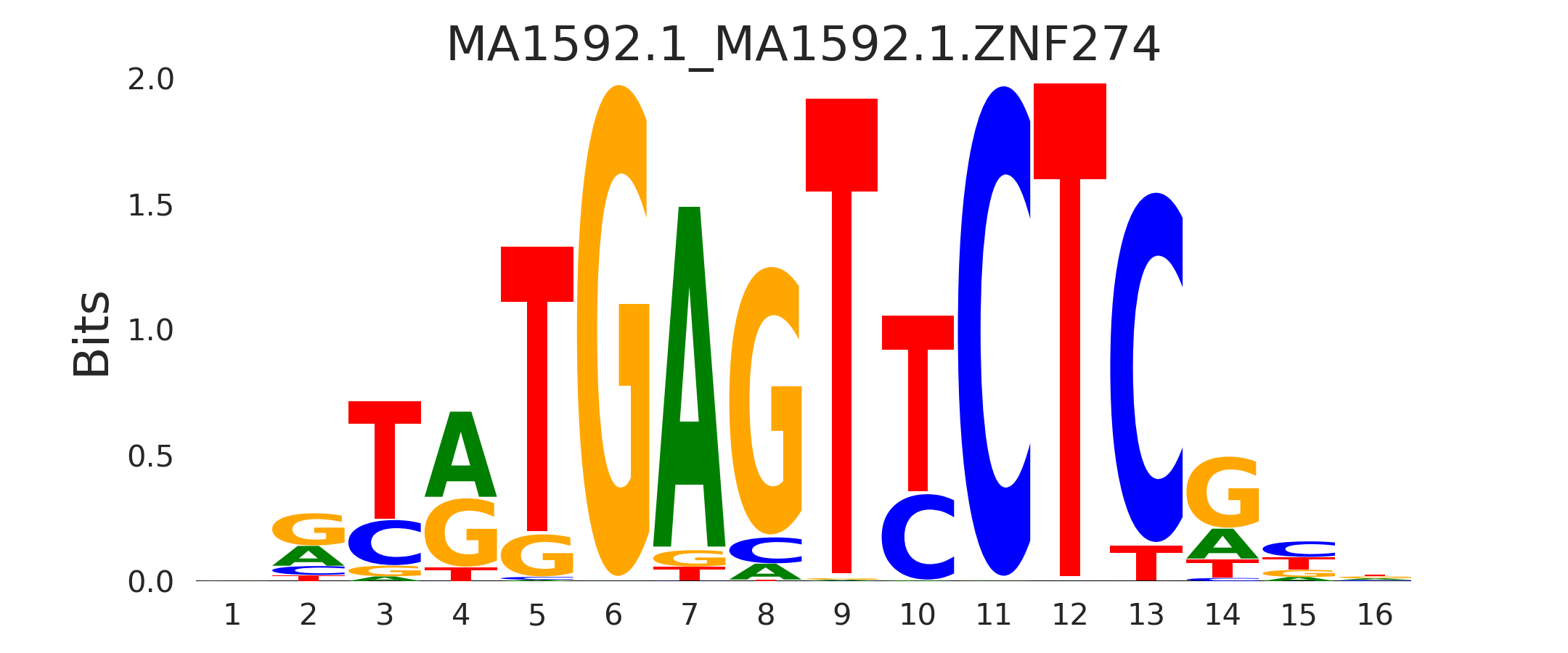
| MA1592.1_MA1592.1.ZNF274 |
MA1592.1.ZNF274,MA0794.1.PROX1 |
|
-1.45 |
-1.26 |
-0.94 |
-1.16 |
3.03 |
-0.57 |
3.36 |
2.92 |
-1.73 |
-1.64 |
-1.42 |
-0.70 |
1.54 |
2.01 |
2.22 |
1.89 |
1 |
0.07 |
0.07 |
0.07 |
0.06 |
-0.02 |
0.01 |
0.01 |
-0.03 |
-0.04 |
-0.05 |
-0.04 |
-0.06 |
-0.02 |
-0.03 |
-0.04 |
-0.03 |
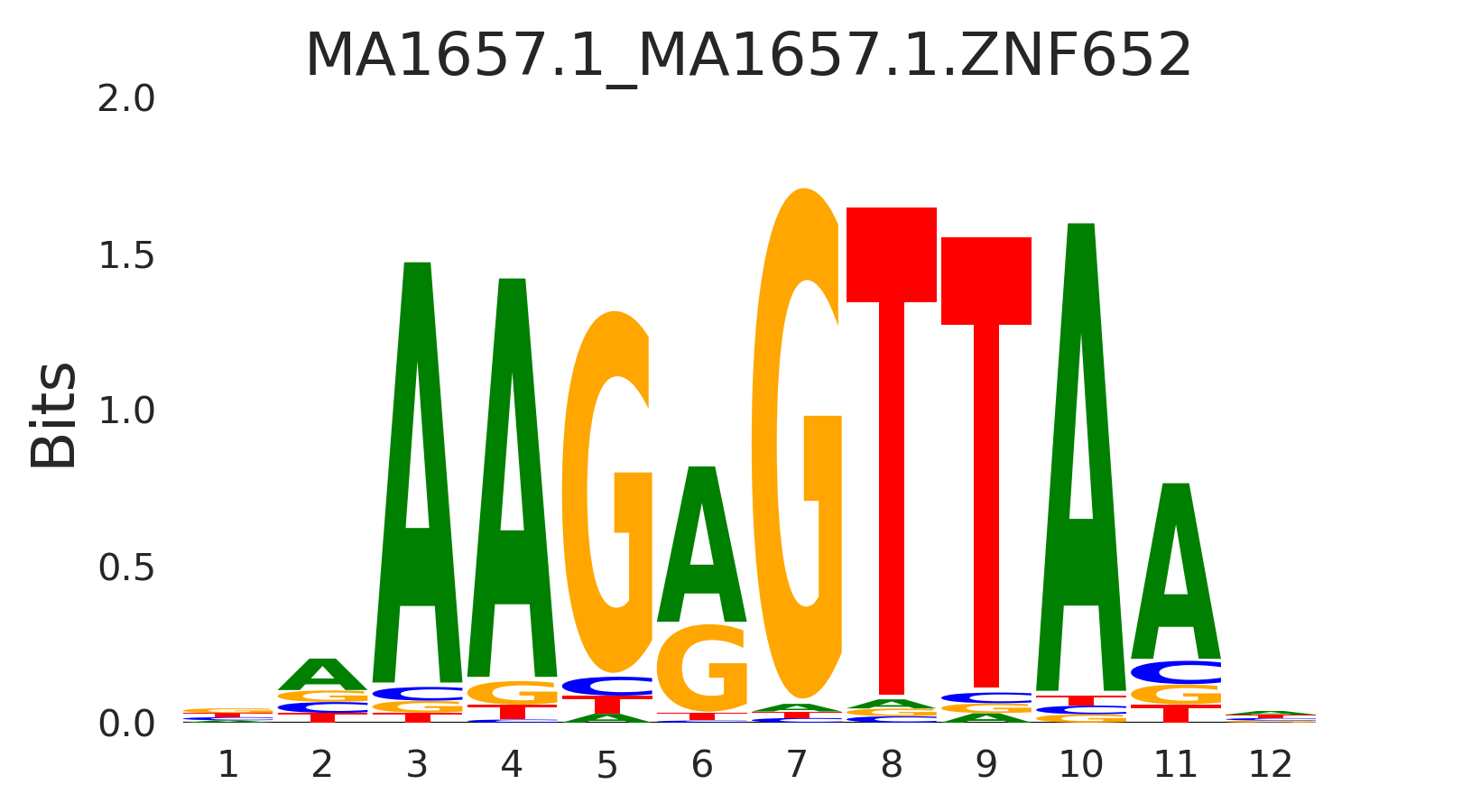
| MA1657.1_MA1657.1.ZNF652 |
MA1657.1.ZNF652,MA0676.1.NR2E1 |
|
-2.74 |
-2.73 |
-2.48 |
-3.02 |
0.20 |
0.20 |
1.30 |
-1.08 |
2.56 |
2.37 |
2.44 |
2.48 |
0.61 |
0.35 |
-0.15 |
0.26 |
1 |
0.05 |
0.05 |
0.06 |
0.04 |
-0.06 |
0.02 |
-0.02 |
-0.07 |
0.04 |
0.03 |
0.03 |
0.00 |
-0.05 |
-0.06 |
-0.09 |
-0.06 |
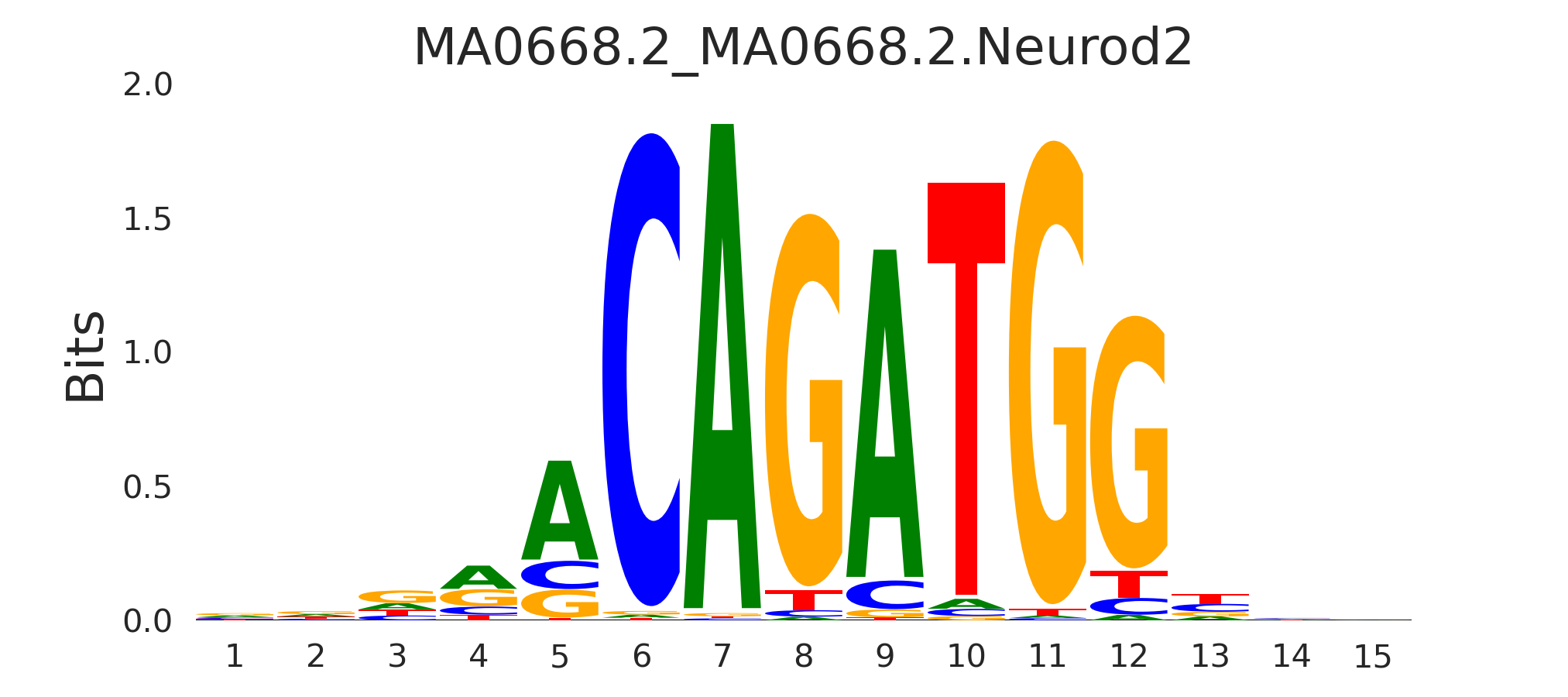
| MA0668.2_MA0668.2.Neurod2 |
MA0668.2.NEUROD2,MA1468.1.ATOH7,MA0827.1.OLIG3,MA1524.2.MSGN1,MA1570.1.TFAP4, (...) |
|
2.11 |
2.08 |
1.81 |
2.30 |
2.42 |
-1.83 |
-1.95 |
2.23 |
-3.54 |
-3.39 |
-3.31 |
-3.42 |
0.62 |
0.98 |
2.11 |
0.76 |
1 |
0.11 |
0.12 |
0.10 |
0.11 |
-0.04 |
-0.01 |
-0.04 |
-0.05 |
-0.06 |
-0.07 |
-0.08 |
-0.08 |
-0.02 |
-0.03 |
-0.03 |
-0.04 |
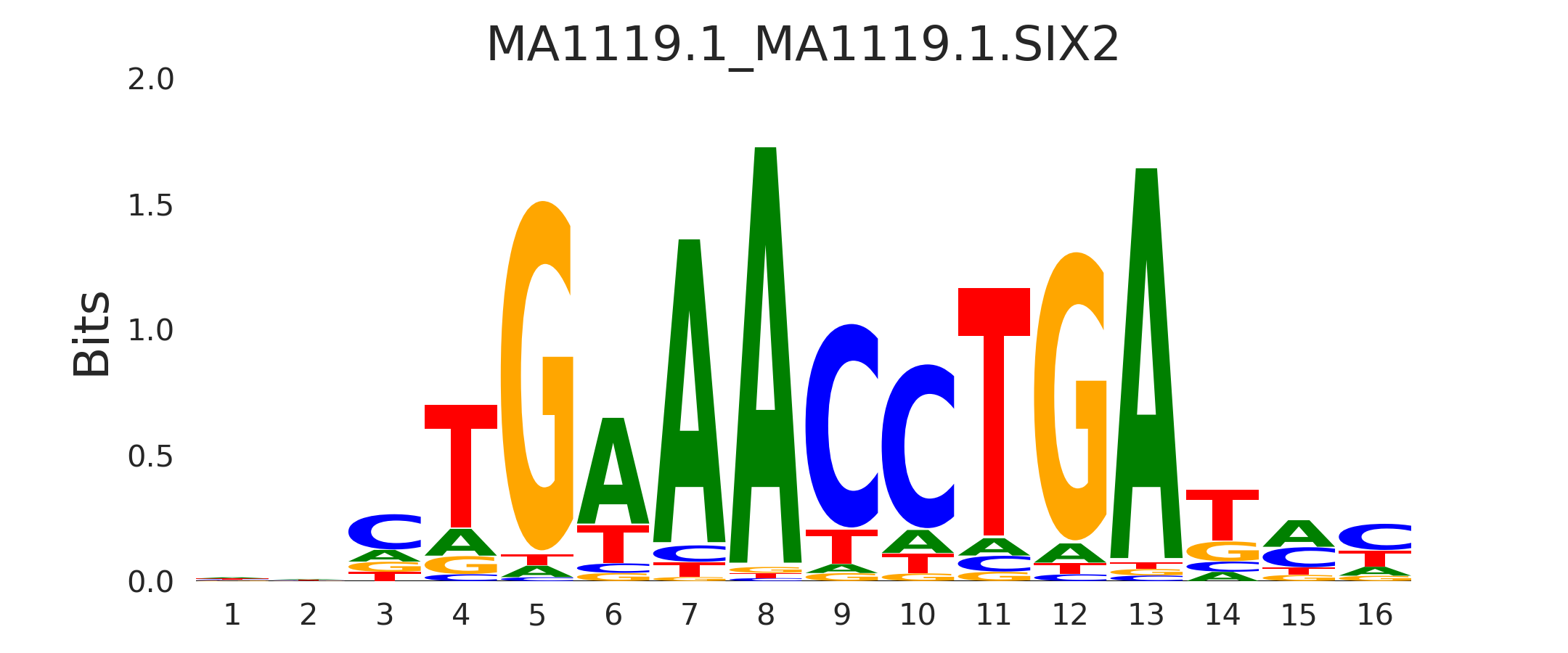
| MA1119.1_MA1119.1.SIX2 |
MA1119.1.SIX2,MA2037.1.SIX4,MA1118.1.SIX1 |
|
-2.30 |
-2.44 |
-2.71 |
-2.40 |
1.16 |
-0.33 |
0.15 |
0.76 |
-1.30 |
-0.80 |
-0.76 |
-0.16 |
3.29 |
3.94 |
2.74 |
3.06 |
1 |
0.04 |
0.04 |
0.04 |
0.04 |
-0.05 |
0.03 |
-0.02 |
-0.06 |
-0.01 |
-0.02 |
-0.02 |
-0.04 |
0.01 |
-0.00 |
-0.04 |
0.01 |
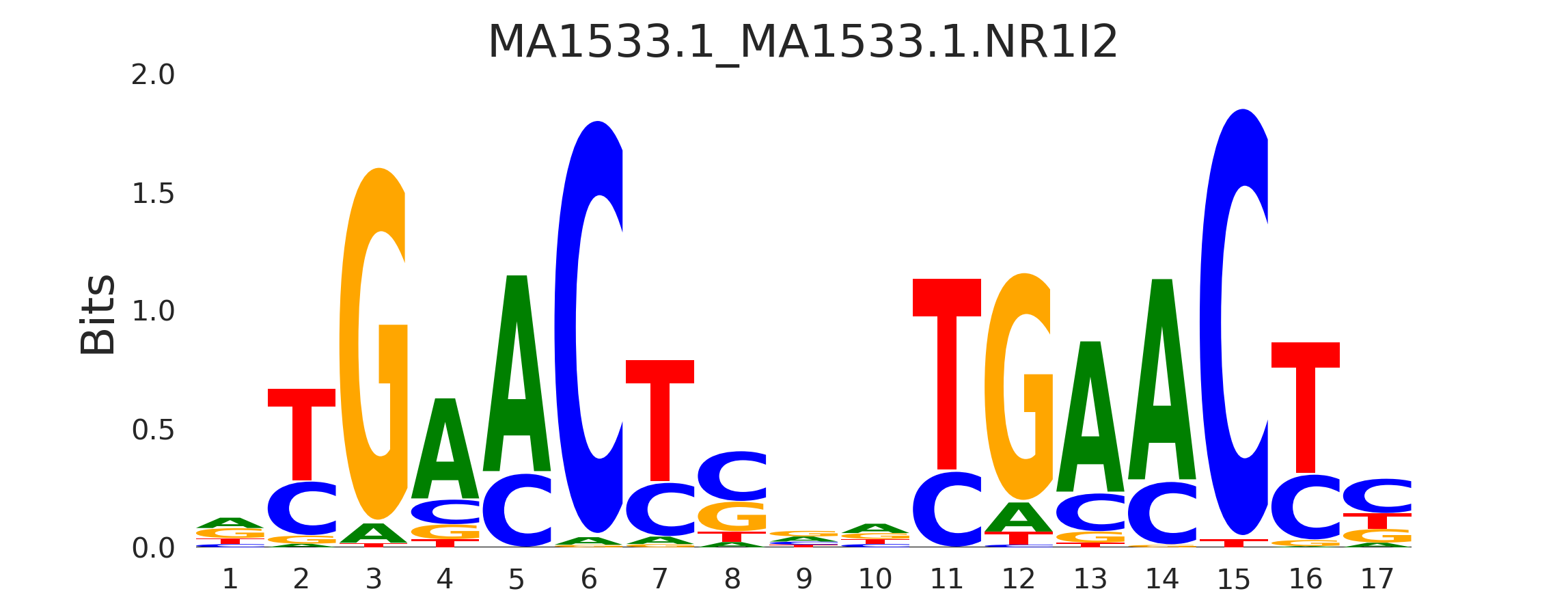
| MA1533.1_MA1533.1.NR1I2 |
MA1533.1.NR1I2,VDR,MA0693.3.VDR,MA1534.1.NR1I3,MA0074.1.RXRA |
|
-2.90 |
-2.95 |
-3.02 |
-2.81 |
0.85 |
0.41 |
-0.57 |
1.17 |
1.76 |
1.96 |
1.67 |
1.68 |
1.17 |
0.86 |
0.46 |
0.58 |
1 |
0.03 |
0.03 |
0.03 |
0.03 |
-0.05 |
0.04 |
-0.03 |
-0.05 |
0.03 |
0.02 |
0.02 |
-0.01 |
-0.02 |
-0.05 |
-0.07 |
-0.04 |
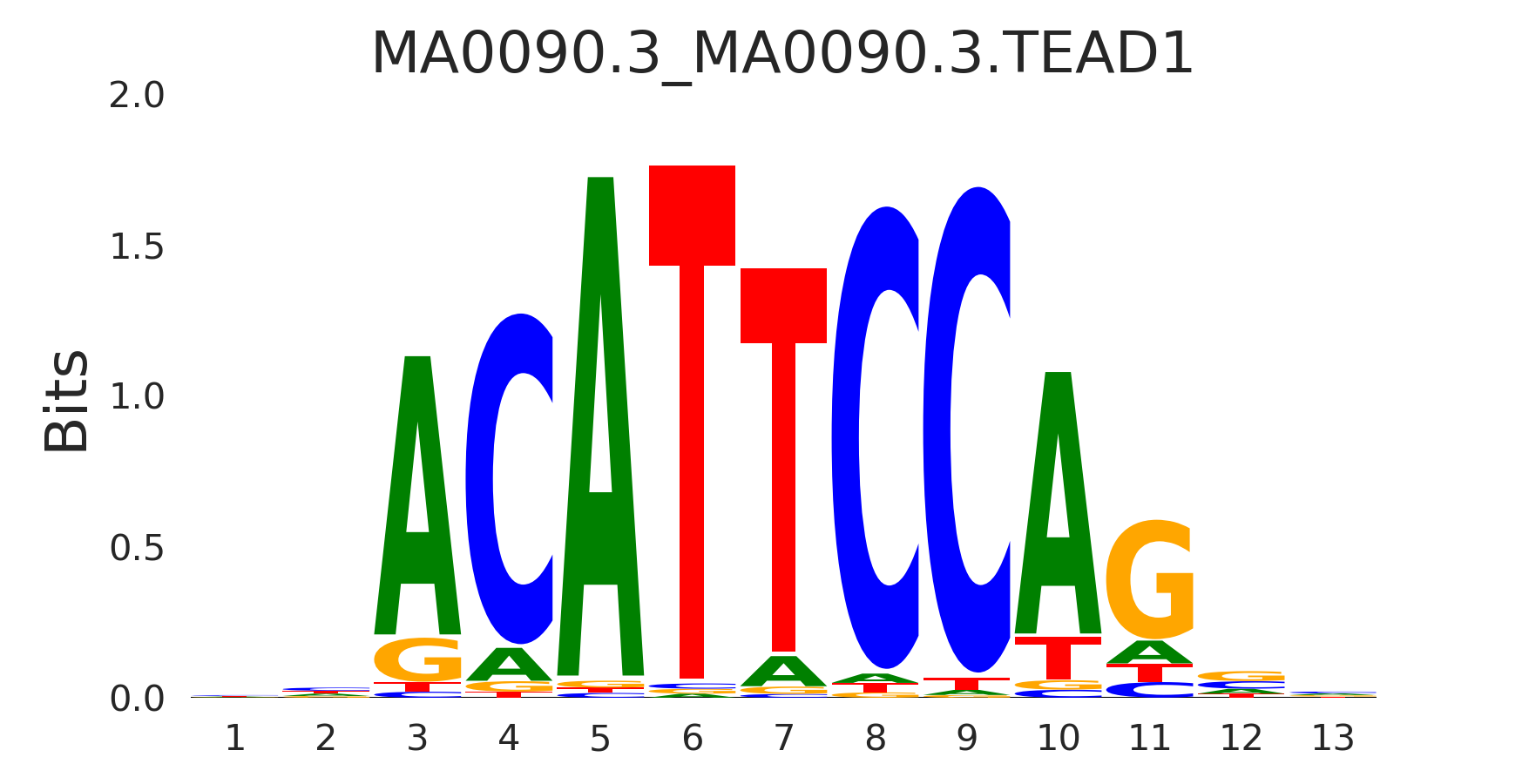
| MA0090.3_MA0090.3.TEAD1 |
MA0090.3.TEAD1,MA0808.1.TEAD3,MA1121.1.TEAD2,MA0809.2.TEAD4 |
|
-1.89 |
-1.33 |
-1.03 |
-1.19 |
3.94 |
0.35 |
3.17 |
1.42 |
-0.71 |
-1.17 |
-0.81 |
-1.25 |
0.81 |
1.05 |
0.81 |
0.45 |
3 |
0.06 |
0.06 |
0.07 |
0.06 |
-0.02 |
0.03 |
0.01 |
-0.05 |
-0.02 |
-0.03 |
-0.02 |
-0.05 |
-0.03 |
-0.04 |
-0.06 |
-0.05 |
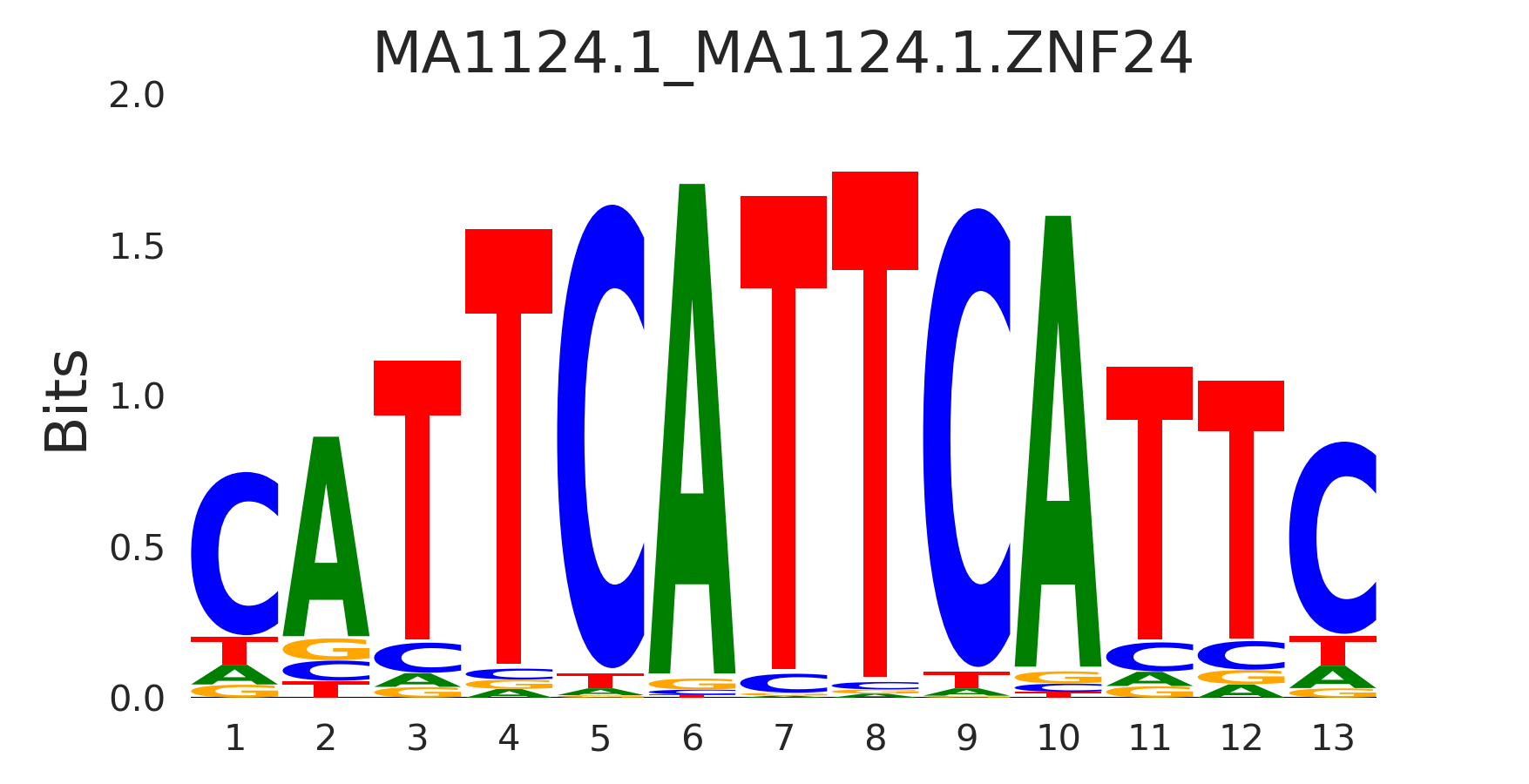
| MA1124.1_MA1124.1.ZNF24 |
MA1124.1.ZNF24,MA1639.1.MEIS1,MA1113.2.PBX2,MA0070.1.PBX1,MA0715.1.PROP1, (...) |
|
3.50 |
3.94 |
3.62 |
4.35 |
-0.62 |
-1.64 |
-1.19 |
-1.14 |
-3.23 |
-3.73 |
-4.03 |
-3.54 |
-1.88 |
-1.61 |
-0.81 |
-0.94 |
1 |
0.18 |
0.18 |
0.17 |
0.17 |
-0.08 |
-0.00 |
-0.04 |
-0.08 |
-0.07 |
-0.08 |
-0.09 |
-0.10 |
-0.08 |
-0.10 |
-0.10 |
-0.09 |
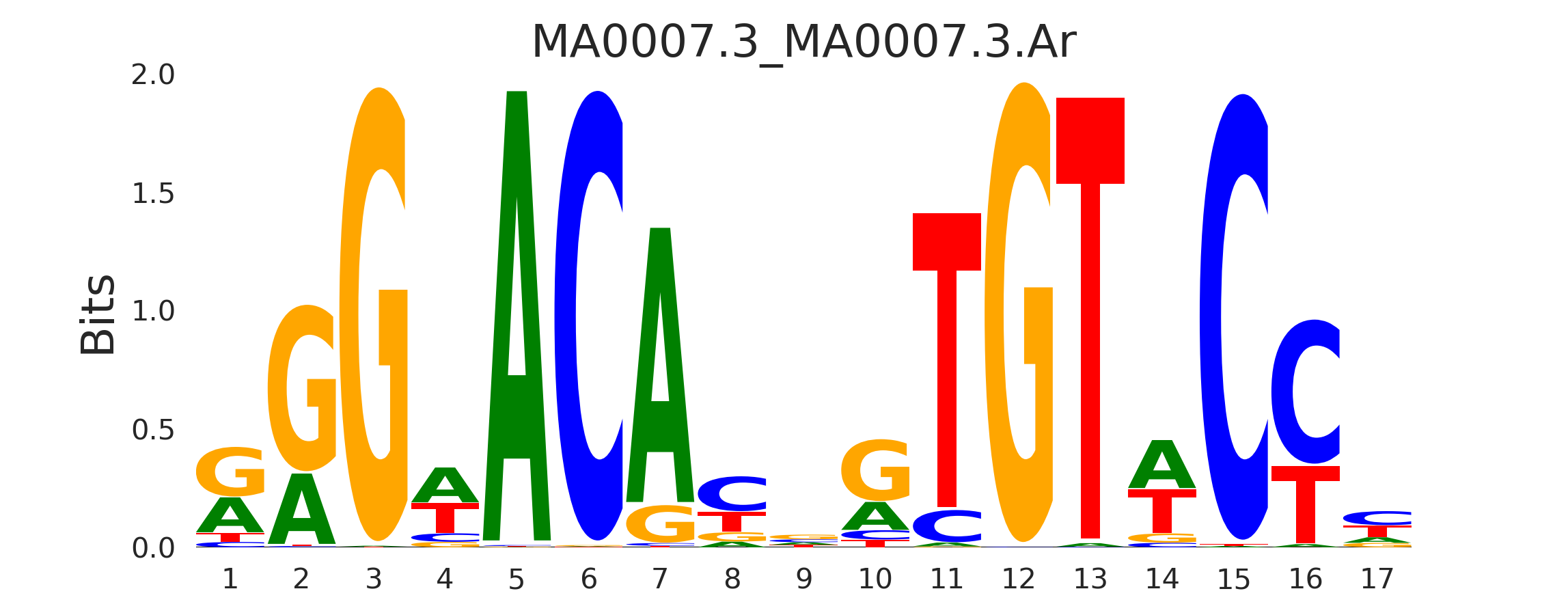
| MA0007.3_MA0007.3.Ar |
MA0007.3.AR,MA0113.3.NR3C1,MA0727.1.NR3C2 |
|
-2.08 |
-2.14 |
-2.11 |
-1.70 |
1.99 |
0.43 |
0.36 |
3.24 |
0.06 |
-0.13 |
-0.41 |
0.93 |
1.93 |
2.11 |
1.65 |
1.83 |
1 |
0.04 |
0.04 |
0.04 |
0.04 |
-0.03 |
0.02 |
-0.01 |
-0.03 |
-0.01 |
-0.02 |
-0.02 |
-0.02 |
-0.01 |
-0.03 |
-0.04 |
-0.03 |
| MA1148.1_MA1148.1.PPARA::RXRA |
RXRA,MA1148.1.PPARA,MA1536.1.NR2C2,MA1574.1.THRB,MA0071.1.RORA, (...) |
|
-3.73 |
-3.73 |
-3.73 |
-3.73 |
0.44 |
-0.42 |
-0.56 |
0.88 |
2.78 |
2.78 |
2.74 |
3.02 |
1.93 |
1.93 |
0.75 |
2.43 |
1 |
-0.01 |
-0.01 |
-0.01 |
-0.02 |
-0.05 |
0.00 |
-0.03 |
-0.05 |
0.07 |
0.06 |
0.06 |
0.05 |
-0.00 |
-0.02 |
-0.05 |
-0.03 |
| MA1868.1_MA1868.1.ZNF320 |
MA1868.1.ZNF320,MA0155.1.INSM1,MA1548.1.PLAGL2,MA0163.1.PLAG1,MA1615.1.PLAGL1 |
|
-1.35 |
-0.96 |
-1.28 |
-1.38 |
1.50 |
-3.23 |
1.73 |
0.05 |
-0.96 |
-1.33 |
-1.23 |
-1.12 |
2.37 |
2.78 |
2.61 |
1.82 |
1 |
0.07 |
0.07 |
0.07 |
0.06 |
-0.02 |
-0.04 |
0.00 |
-0.04 |
-0.06 |
-0.06 |
-0.06 |
-0.07 |
0.02 |
0.01 |
-0.00 |
-0.01 |
| MA0153.2_MA0153.2.HNF1B |
MA0153.2.HNF1B,MA0125.1.NOBOX,MA0882.1.DLX6,MA0889.1.GBX1,MA0661.1.MEOX1, (...) |
|
-1.14 |
-1.22 |
-2.37 |
-1.56 |
-3.01 |
-2.56 |
-3.14 |
-2.69 |
3.14 |
3.62 |
3.39 |
3.54 |
-1.98 |
-2.11 |
-1.80 |
-1.90 |
3 |
0.11 |
0.10 |
0.09 |
0.10 |
-0.13 |
0.02 |
-0.09 |
-0.12 |
0.08 |
0.07 |
0.06 |
0.04 |
-0.12 |
-0.13 |
-0.16 |
-0.12 |
| MA0480.2_MA0480.2.Foxo1 |
MA0480.2.FOXO1,MA0613.1.FOXG1,MA1683.1.FOXA3,MA0033.2.FOXL1,MA0040.1.FOXQ1, (...) |
|
-1.15 |
-1.30 |
-1.80 |
-1.63 |
-2.09 |
-1.82 |
-0.97 |
-3.14 |
2.95 |
3.08 |
3.14 |
2.74 |
-1.11 |
-1.51 |
-2.00 |
-2.39 |
2 |
0.07 |
0.07 |
0.07 |
0.06 |
-0.10 |
0.01 |
-0.03 |
-0.11 |
0.07 |
0.06 |
0.06 |
0.02 |
-0.08 |
-0.09 |
-0.13 |
-0.10 |
| MA1509.1_MA1509.1.IRF6 |
MA1509.1.IRF6,MA2020.1.ZBED2,MA1647.2.PRDM4,MA1420.1.IRF5 |
|
-1.40 |
-1.66 |
-0.90 |
-1.27 |
-0.91 |
3.31 |
2.56 |
-0.70 |
2.24 |
2.37 |
2.44 |
2.30 |
-1.96 |
-2.00 |
-1.97 |
-1.34 |
4 |
0.08 |
0.07 |
0.09 |
0.08 |
-0.09 |
0.10 |
-0.00 |
-0.08 |
0.05 |
0.03 |
0.04 |
-0.00 |
-0.12 |
-0.13 |
-0.18 |
-0.11 |
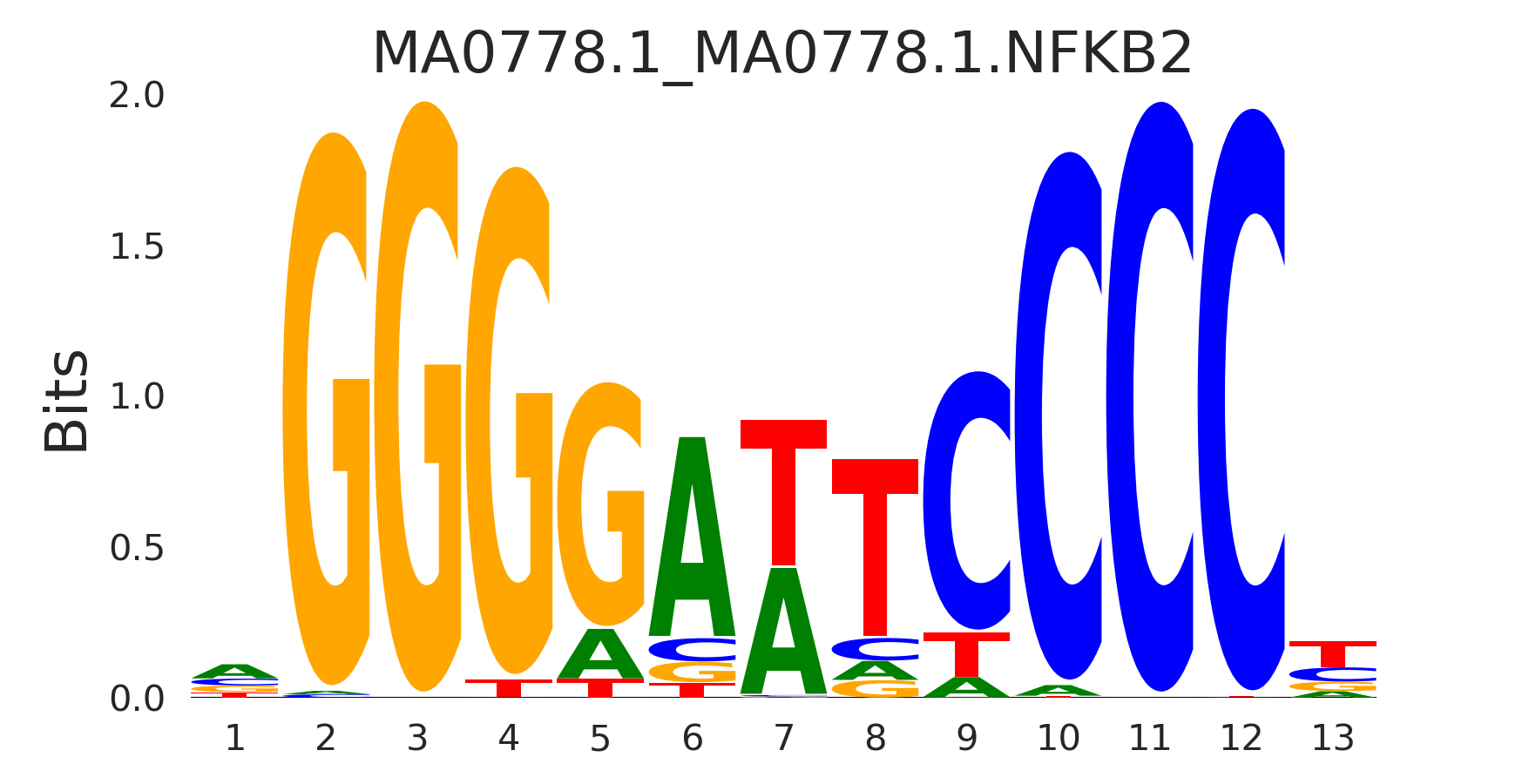
| MA0778.1_MA0778.1.NFKB2 |
MA0778.1.NFKB2,MA0056.2.MZF1,MA0101.1.REL,MA0105.4.NFKB1,MA1117.1.RELB, (...) |
|
-2.83 |
-2.51 |
-2.23 |
-2.89 |
3.50 |
1.51 |
2.97 |
4.35 |
-1.60 |
-1.84 |
-1.12 |
-0.39 |
2.81 |
2.60 |
3.39 |
2.74 |
2 |
0.05 |
0.05 |
0.06 |
0.04 |
-0.02 |
0.04 |
0.01 |
-0.01 |
-0.04 |
-0.05 |
-0.04 |
-0.06 |
-0.01 |
-0.03 |
-0.04 |
-0.03 |
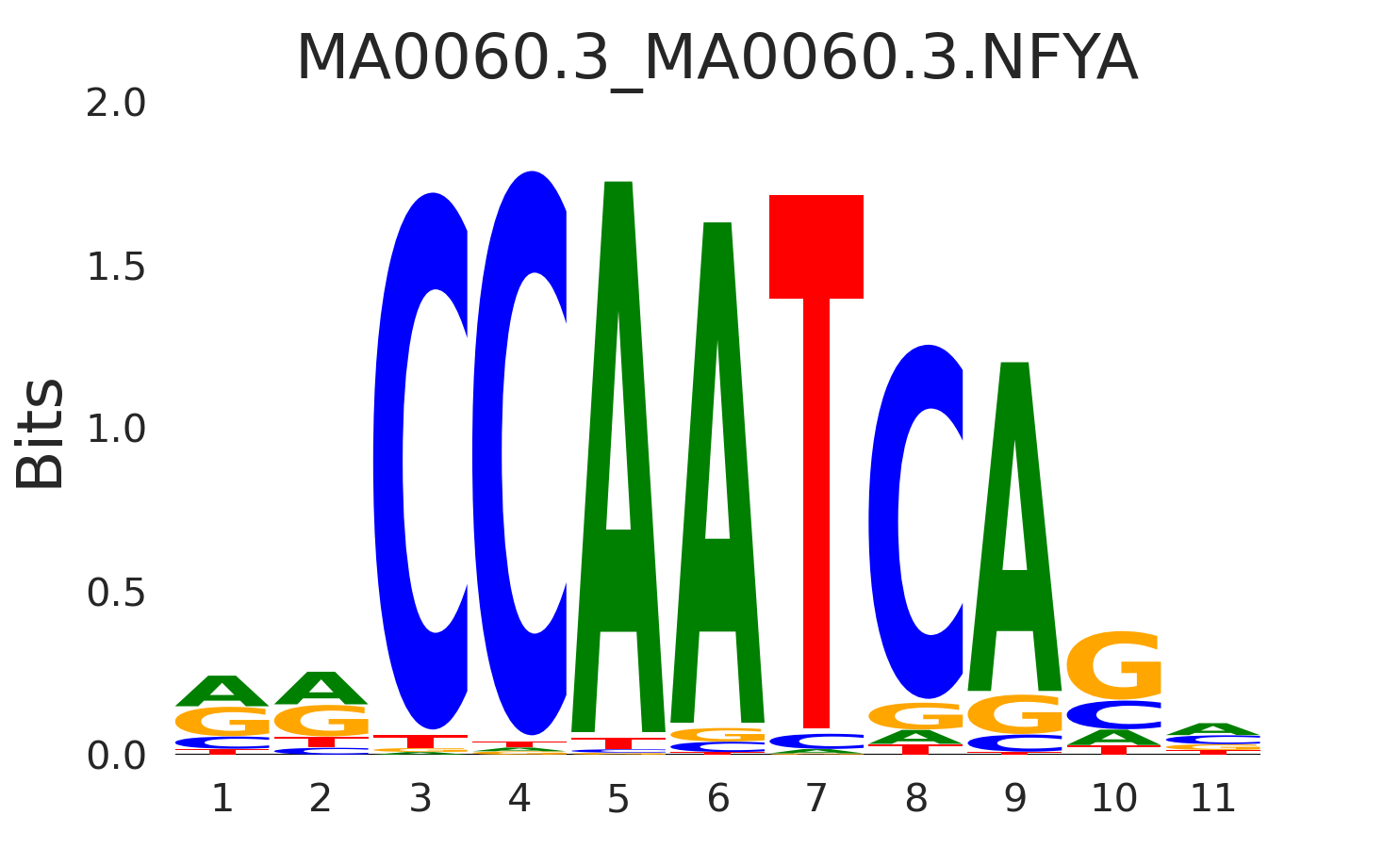
| MA0060.3_MA0060.3.NFYA |
MA0060.3.NFYA,MA0483.1.GFI1B,MA0502.2.NFYB,MA0038.2.GFI1,MA1644.1.NFYC |
|
1.75 |
1.69 |
0.22 |
1.26 |
-1.59 |
1.58 |
-4.35 |
-1.61 |
2.28 |
2.56 |
1.88 |
2.44 |
-2.23 |
-2.09 |
-2.56 |
-2.23 |
3 |
0.11 |
0.11 |
0.09 |
0.10 |
-0.08 |
0.05 |
-0.11 |
-0.08 |
0.02 |
0.02 |
0.01 |
-0.01 |
-0.09 |
-0.11 |
-0.14 |
-0.09 |
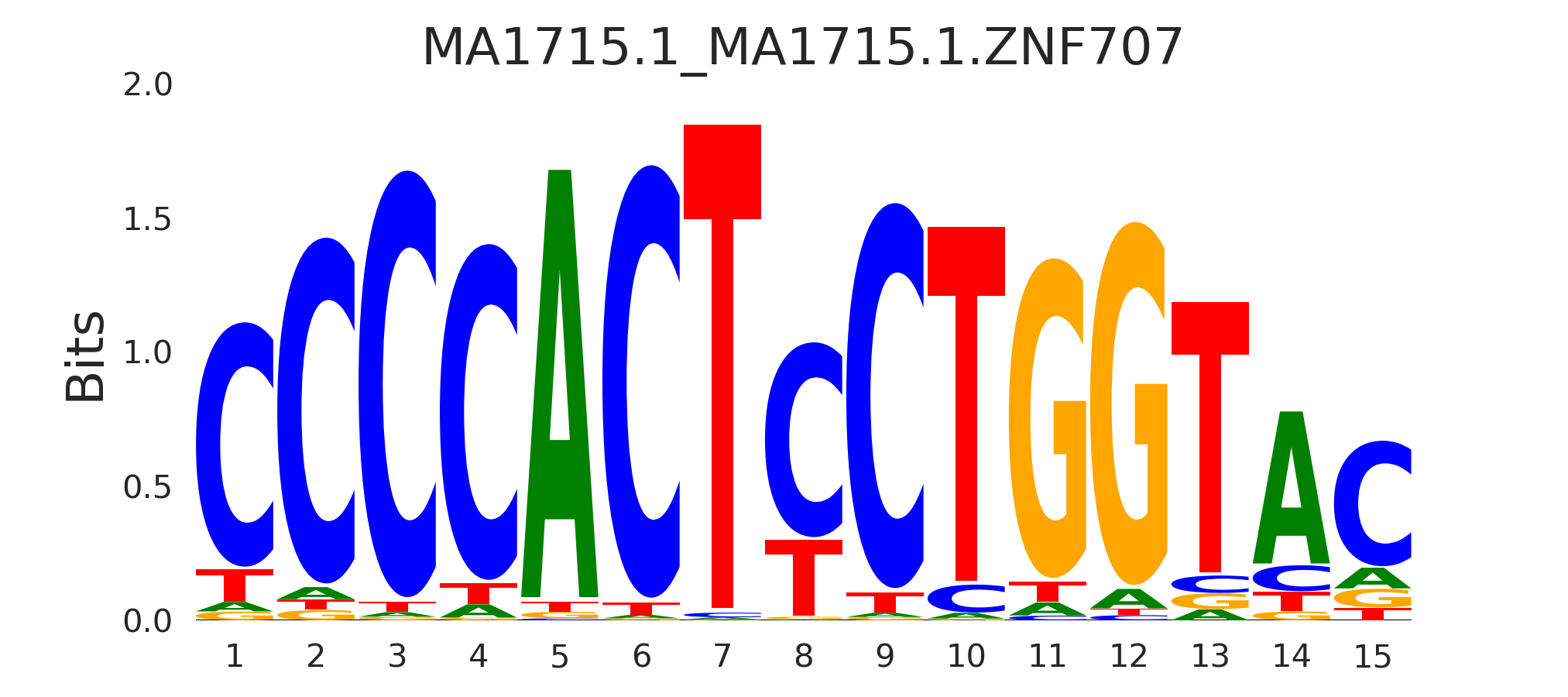
| MA1715.1_MA1715.1.ZNF707 |
MA1715.1.ZNF707 |
|
-1.34 |
-1.38 |
-1.57 |
-1.19 |
2.05 |
-0.49 |
1.00 |
3.10 |
-1.08 |
-1.43 |
-1.06 |
-0.41 |
1.46 |
2.22 |
2.27 |
1.96 |
1 |
0.07 |
0.07 |
0.07 |
0.06 |
-0.04 |
0.00 |
-0.01 |
-0.04 |
-0.03 |
-0.04 |
-0.04 |
-0.05 |
-0.01 |
-0.03 |
-0.04 |
-0.03 |
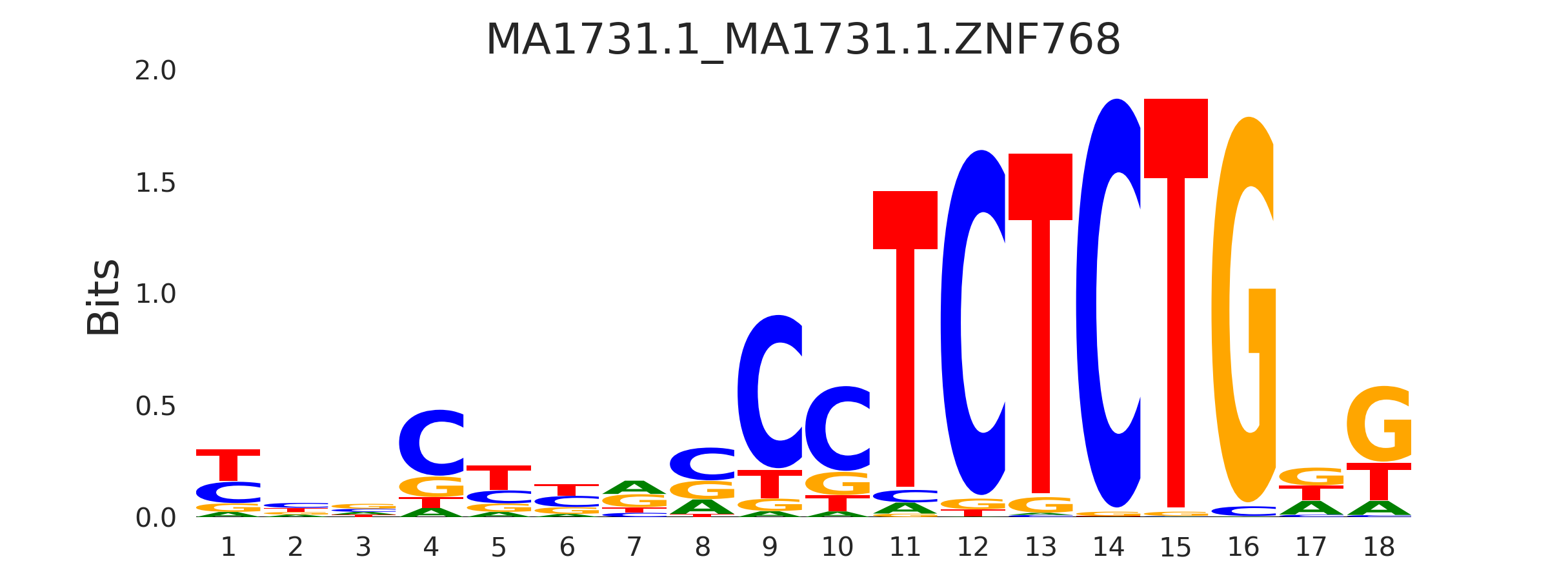
| MA1731.1_MA1731.1.ZNF768 |
MA1731.1.ZNF768,MA2032.1.SP5,MA1627.1.WT1,MA0149.1.EWSR1-FLI1,MA1652.1.ZKSCAN5, (...) |
|
0.92 |
1.77 |
1.77 |
1.30 |
2.74 |
-4.35 |
1.60 |
1.64 |
-4.35 |
-4.35 |
-3.34 |
-2.78 |
3.42 |
3.10 |
4.35 |
2.89 |
1 |
0.08 |
0.10 |
0.10 |
0.08 |
-0.00 |
-0.10 |
0.02 |
-0.03 |
-0.11 |
-0.11 |
-0.10 |
-0.09 |
0.04 |
0.03 |
0.05 |
0.01 |
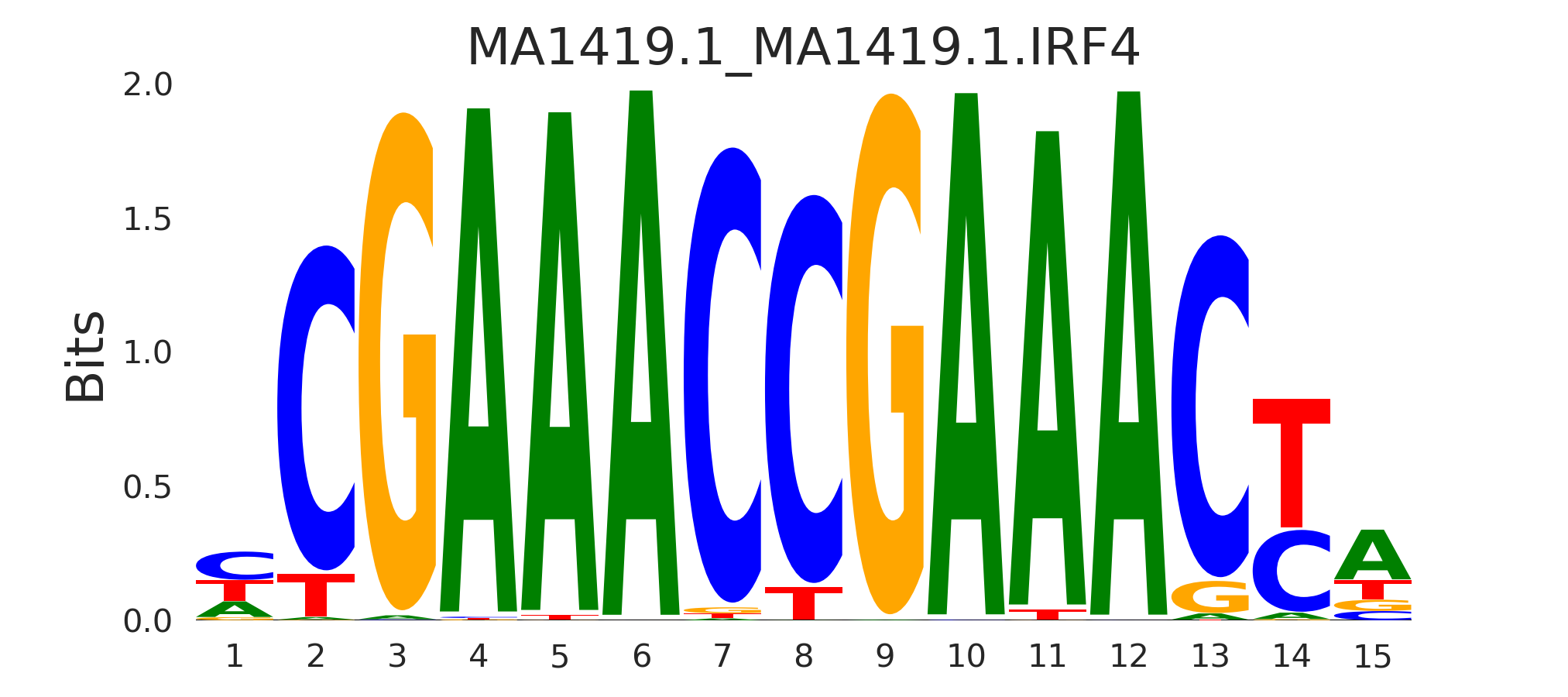
| MA1419.1_MA1419.1.IRF4 |
MA1419.1.IRF4,STAT2,MA0653.1.IRF9,MA0051.1.IRF2,MA0652.1.IRF8, (...) |
|
-2.10 |
-1.95 |
-1.45 |
-2.17 |
-0.34 |
1.27 |
4.03 |
-0.21 |
2.52 |
2.47 |
2.69 |
2.74 |
-1.52 |
-1.83 |
-1.65 |
-1.21 |
3 |
0.07 |
0.06 |
0.08 |
0.06 |
-0.08 |
0.07 |
0.01 |
-0.08 |
0.05 |
0.04 |
0.06 |
0.01 |
-0.11 |
-0.12 |
-0.16 |
-0.10 |
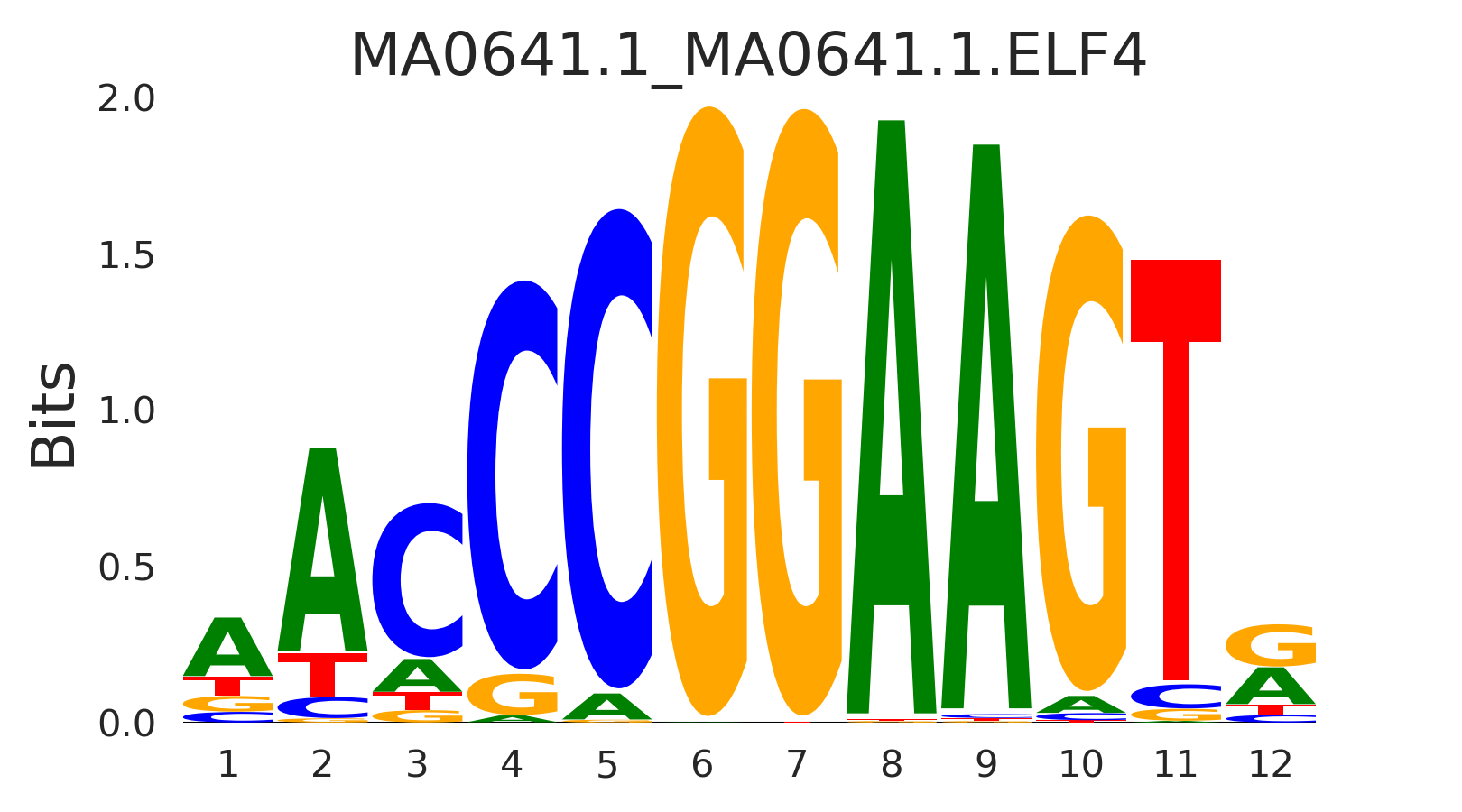
| MA0641.1_MA0641.1.ELF4 |
DRGX,HOXB13,MA0641.1.ELF4,MA1793.1.ETV5,SREBF2, (...) |
|
0.68 |
-0.86 |
-0.11 |
-0.11 |
-2.89 |
1.59 |
-0.29 |
-3.39 |
3.82 |
3.50 |
3.73 |
3.10 |
-3.95 |
-4.35 |
-4.35 |
-3.73 |
1 |
0.12 |
0.10 |
0.11 |
0.10 |
-0.11 |
0.05 |
-0.03 |
-0.13 |
0.10 |
0.07 |
0.09 |
0.01 |
-0.15 |
-0.18 |
-0.23 |
-0.16 |
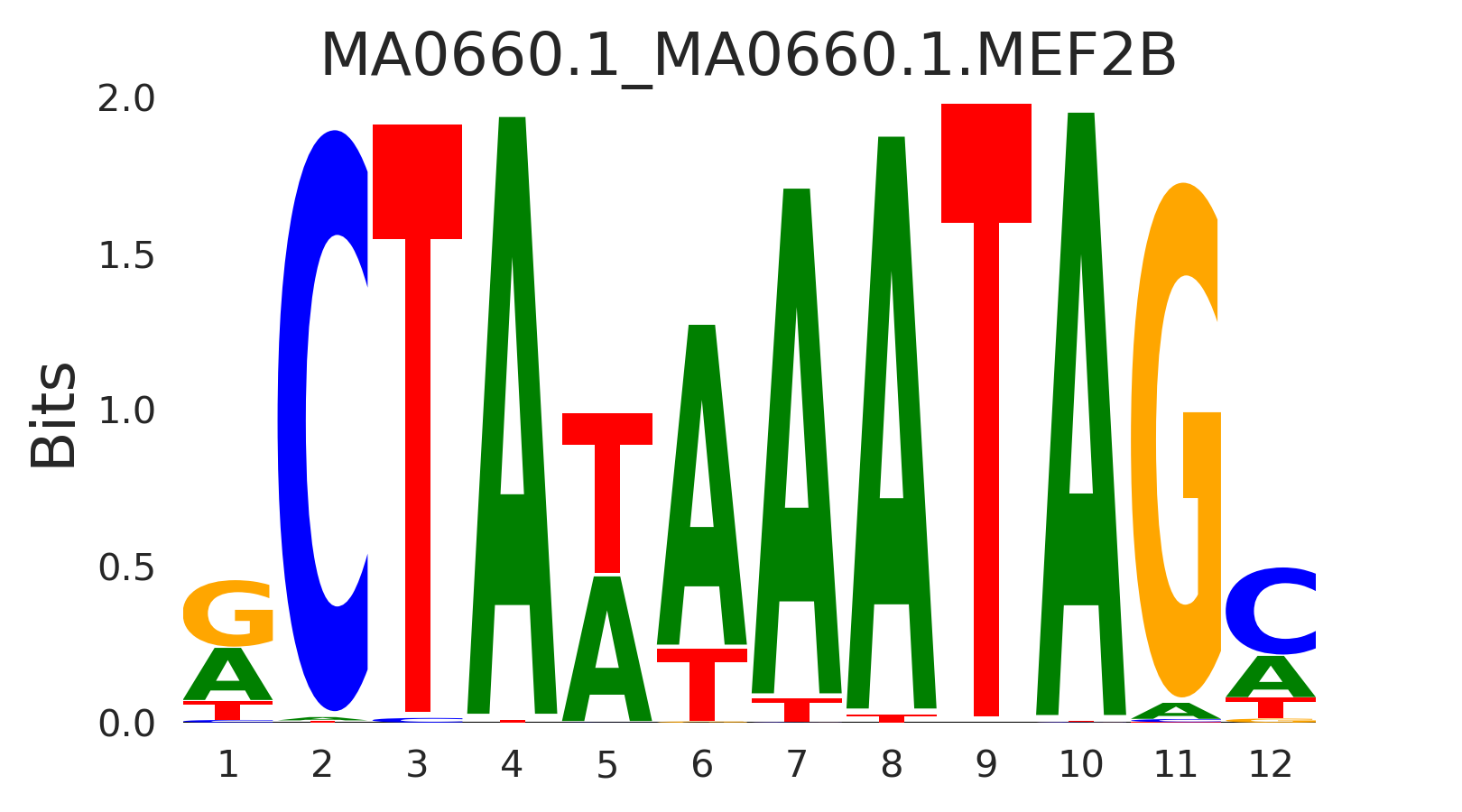
| MA0660.1_MA0660.1.MEF2B |
MA0660.1.MEF2B,MA0052.4.MEF2A,MA0465.2.CDX2,MA0901.2.HOXB13,MA0497.1.MEF2C, (...) |
|
1.32 |
1.02 |
0.22 |
1.13 |
-3.24 |
-2.09 |
-3.50 |
-2.66 |
-2.45 |
-2.44 |
-2.50 |
-3.16 |
4.03 |
3.78 |
2.90 |
4.15 |
3 |
0.12 |
0.12 |
0.11 |
0.11 |
-0.13 |
0.01 |
-0.08 |
-0.11 |
-0.05 |
-0.06 |
-0.06 |
-0.09 |
0.01 |
-0.01 |
-0.06 |
0.01 |
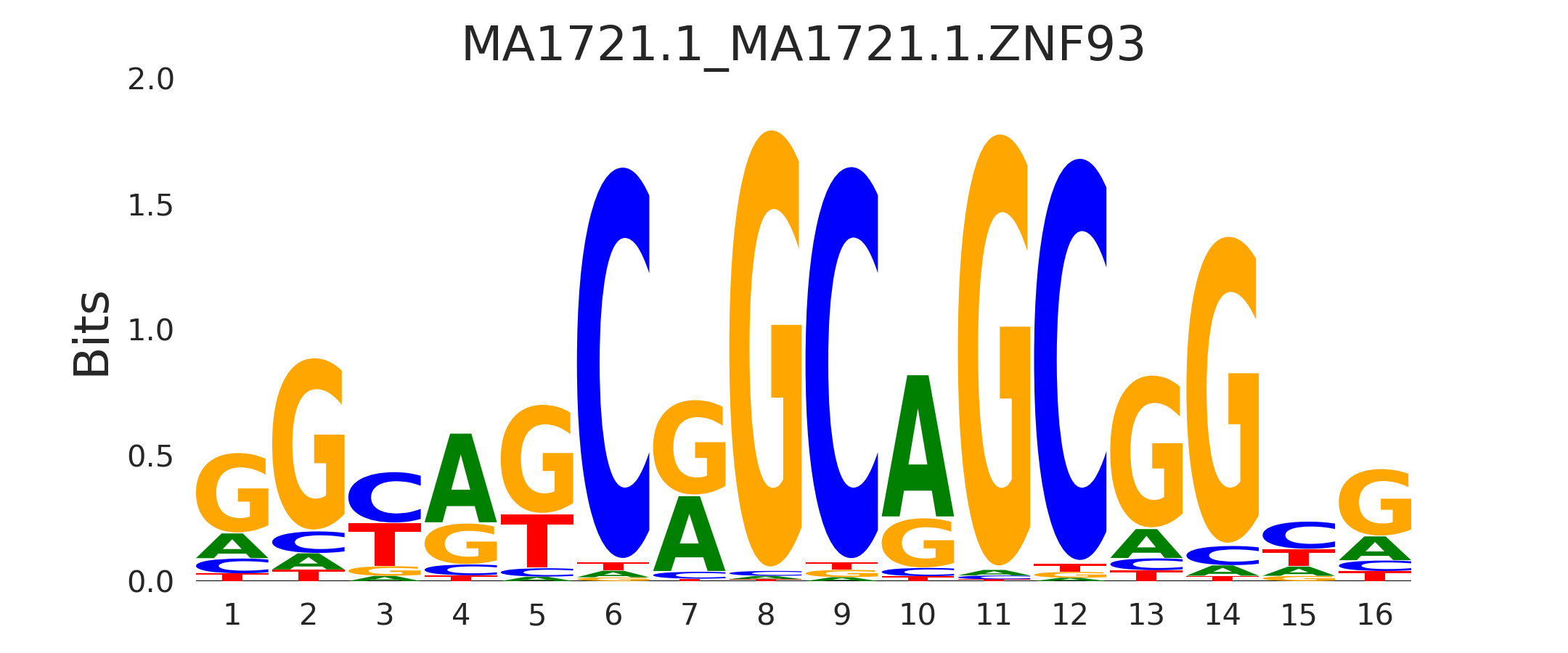
| MA1721.1_MA1721.1.ZNF93 |
MA1721.1.ZNF93,MA1797.1.ERF,NHLH1,MA1713.1.ZNF610,MA1583.1.ZFP57 |
|
2.40 |
2.56 |
3.15 |
2.14 |
-1.46 |
-1.57 |
-0.03 |
-0.19 |
-3.50 |
-3.08 |
-3.94 |
-4.35 |
0.96 |
0.37 |
0.74 |
1.53 |
2 |
0.16 |
0.16 |
0.17 |
0.15 |
-0.09 |
0.01 |
-0.02 |
-0.08 |
-0.09 |
-0.09 |
-0.10 |
-0.12 |
-0.05 |
-0.06 |
-0.08 |
-0.06 |
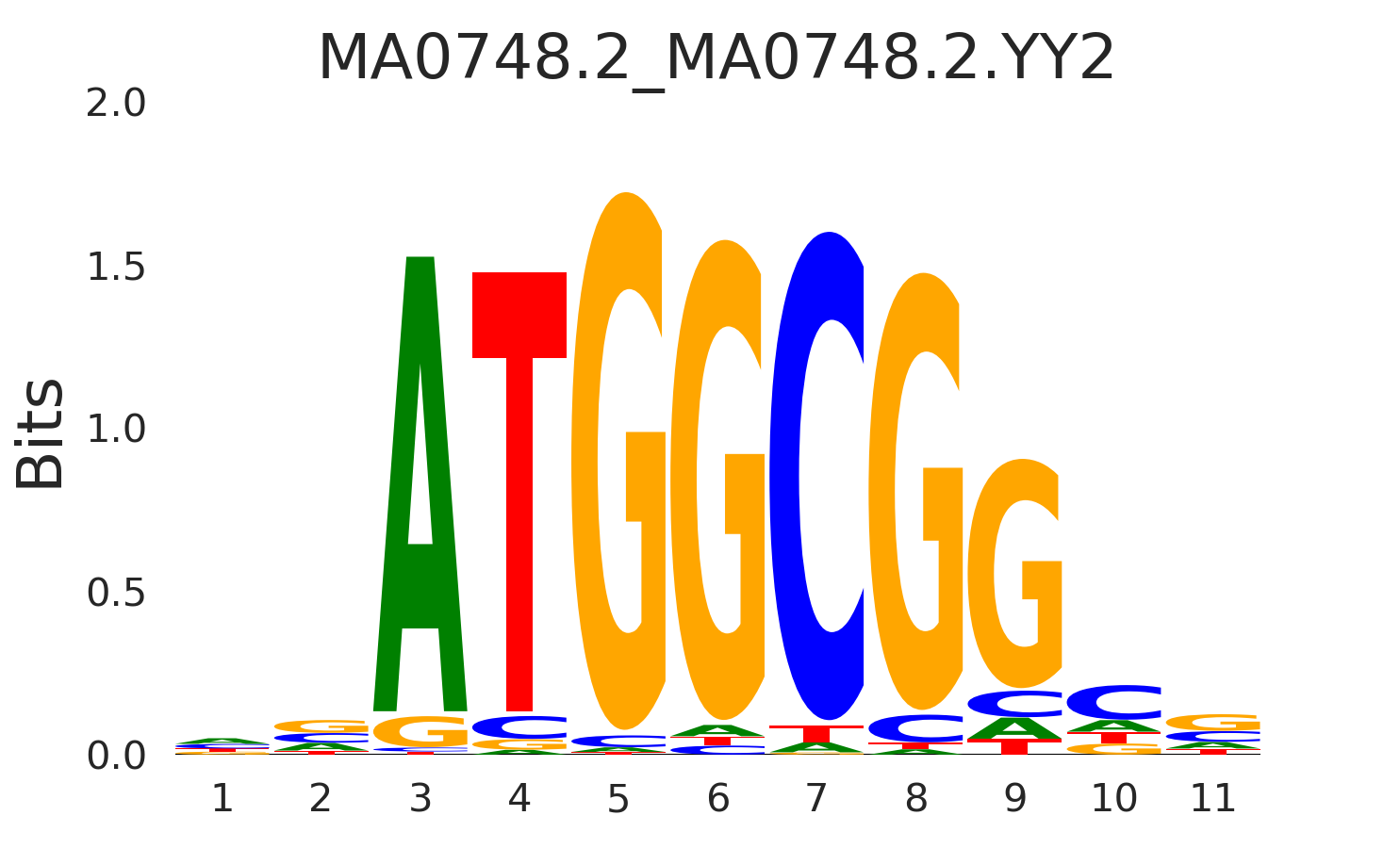
| MA0748.2_MA0748.2.YY2 |
MA0748.2.YY2,MA0469.3.E2F3,MA1651.1.ZFP42,MA0024.3.E2F1,MA0864.2.E2F2, (...) |
|
3.62 |
3.39 |
3.24 |
3.39 |
-3.73 |
0.35 |
-2.70 |
-3.73 |
2.35 |
1.74 |
0.72 |
-0.48 |
-3.10 |
-3.50 |
-3.39 |
-3.39 |
3 |
0.21 |
0.19 |
0.19 |
0.19 |
-0.15 |
0.06 |
-0.09 |
-0.15 |
0.03 |
0.00 |
-0.01 |
-0.06 |
-0.15 |
-0.18 |
-0.23 |
-0.16 |
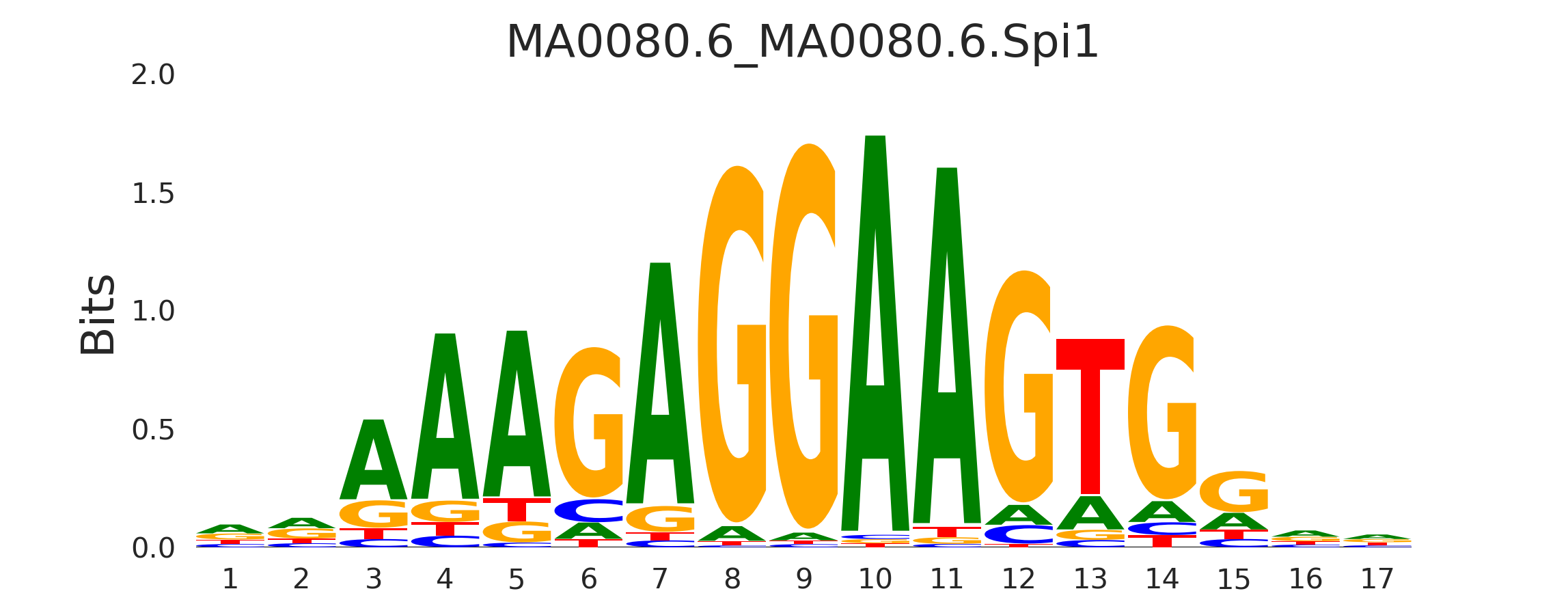
| MA0080.6_MA0080.6.Spi1 |
FOXI1,ELF1,FOXO1,MA0080.6.SPI1,MA1784.1.ERF, (...) |
|
2.03 |
2.17 |
2.15 |
1.97 |
0.98 |
-3.73 |
3.22 |
0.17 |
-2.48 |
-2.80 |
-1.81 |
-2.45 |
0.18 |
-0.19 |
0.64 |
-0.38 |
2 |
0.13 |
0.13 |
0.14 |
0.12 |
-0.04 |
-0.07 |
0.02 |
-0.07 |
-0.05 |
-0.07 |
-0.04 |
-0.08 |
-0.05 |
-0.06 |
-0.06 |
-0.07 |
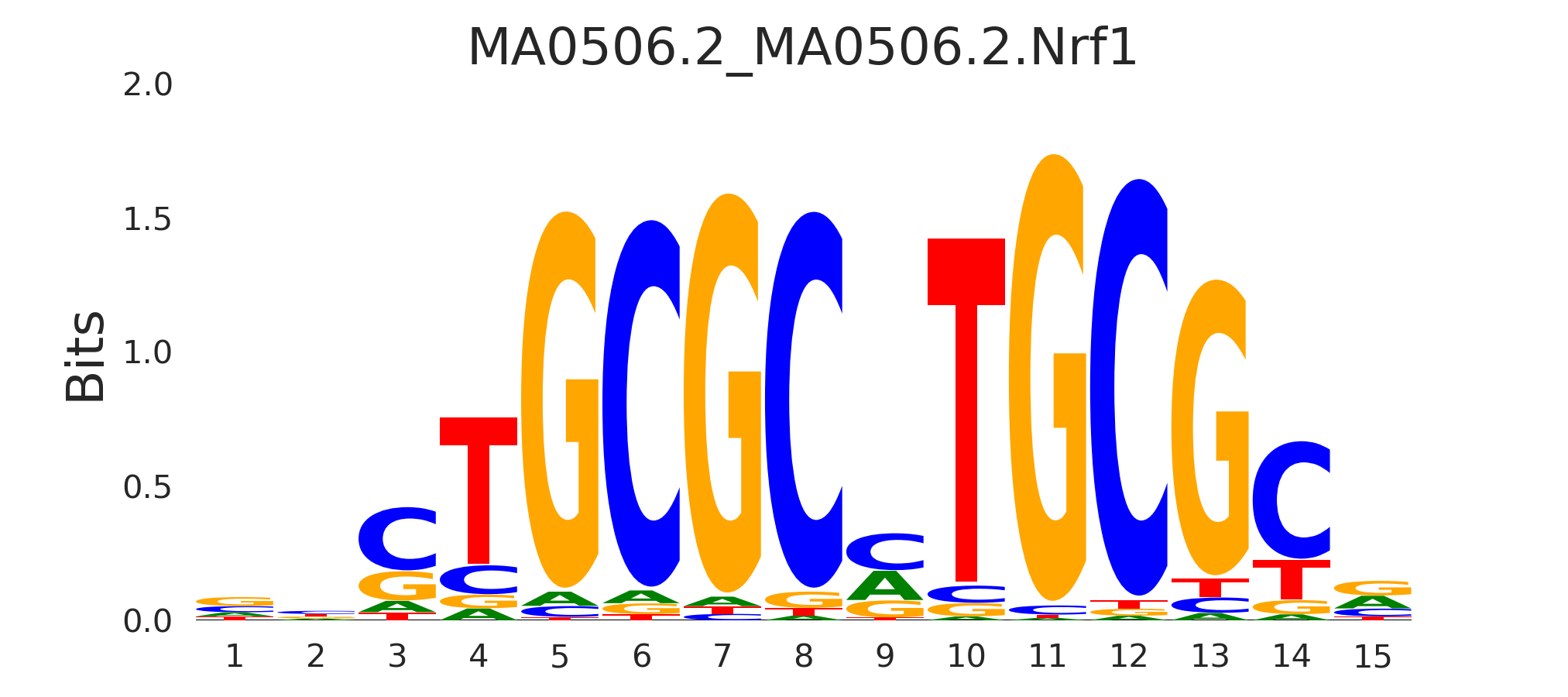
| MA0506.2_MA0506.2.Nrf1 |
MA0506.2.NRF1,MA0472.2.EGR2,ARNT,MA0733.1.EGR4,MA0632.2.TCFL5, (...) |
|
2.64 |
2.78 |
2.48 |
3.01 |
-1.66 |
-0.29 |
-2.99 |
-2.00 |
-1.69 |
-1.85 |
-1.55 |
-1.18 |
-1.70 |
-1.20 |
-1.15 |
-1.52 |
3 |
0.15 |
0.16 |
0.15 |
0.15 |
-0.09 |
0.03 |
-0.07 |
-0.08 |
-0.05 |
-0.06 |
-0.06 |
-0.07 |
-0.08 |
-0.09 |
-0.11 |
-0.09 |
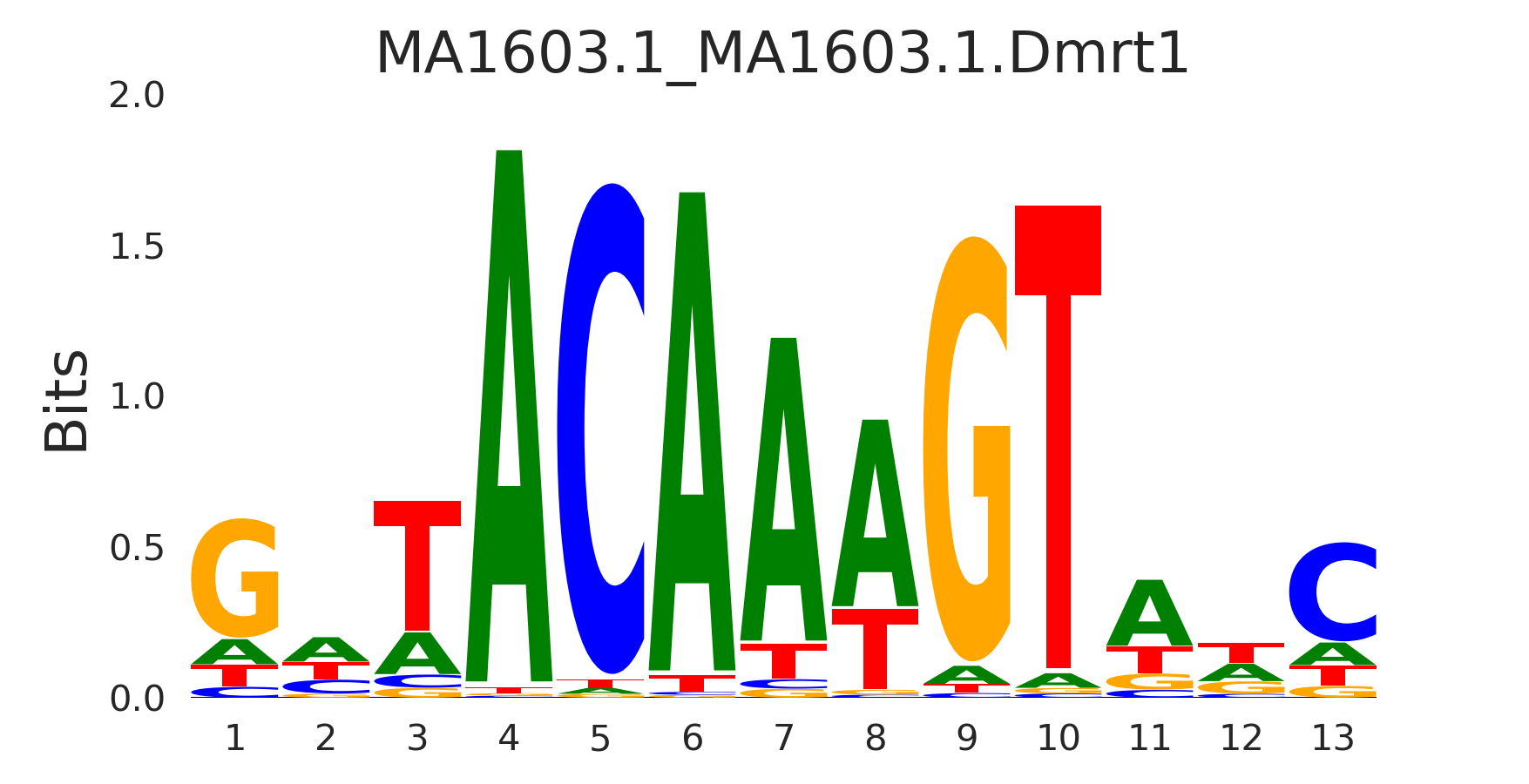
| MA1603.1_MA1603.1.Dmrt1 |
MA1603.1.DMRT1,MA0610.1.DMRT3,MA1707.1.DMRTA1,MA1479.1.DMRTC2,MA1478.1.DMRTA2 |
|
-0.63 |
-0.65 |
-0.50 |
-0.55 |
-1.53 |
4.35 |
2.96 |
0.88 |
1.96 |
2.03 |
2.13 |
1.16 |
-1.92 |
-1.95 |
-2.78 |
-2.40 |
4 |
0.09 |
0.08 |
0.09 |
0.08 |
-0.09 |
0.09 |
-0.01 |
-0.06 |
0.03 |
0.02 |
0.03 |
-0.02 |
-0.10 |
-0.12 |
-0.17 |
-0.11 |
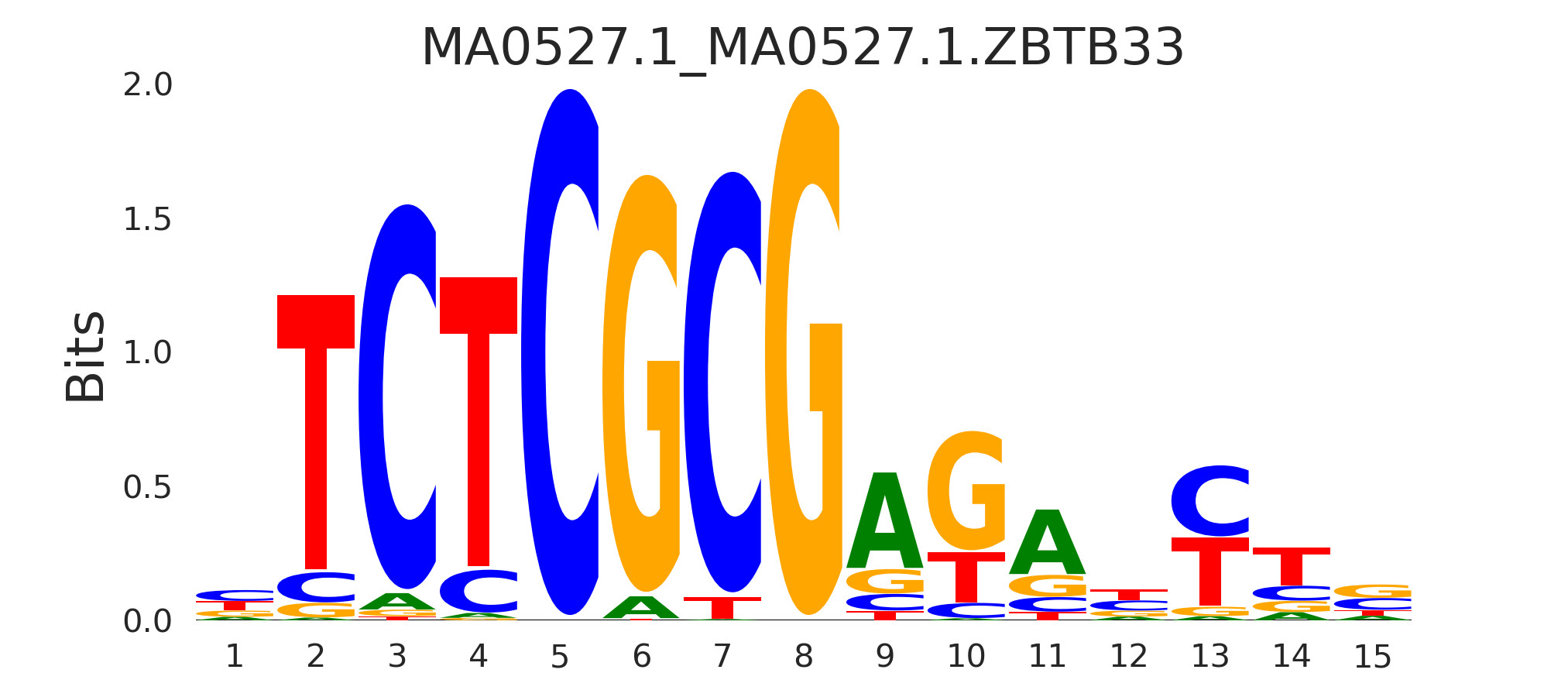
| MA0527.1_MA0527.1.ZBTB33 |
MA0527.1.ZBTB33,MA0749.1.ZBED1,MA2042.1.NPAS4 |
|
4.35 |
4.15 |
4.35 |
3.73 |
-2.48 |
3.02 |
-1.34 |
-1.95 |
-1.88 |
-1.53 |
-1.63 |
-2.70 |
-3.24 |
-3.01 |
-2.80 |
-2.67 |
4 |
0.19 |
0.19 |
0.20 |
0.18 |
-0.11 |
0.08 |
-0.05 |
-0.10 |
-0.04 |
-0.05 |
-0.05 |
-0.10 |
-0.14 |
-0.15 |
-0.19 |
-0.14 |
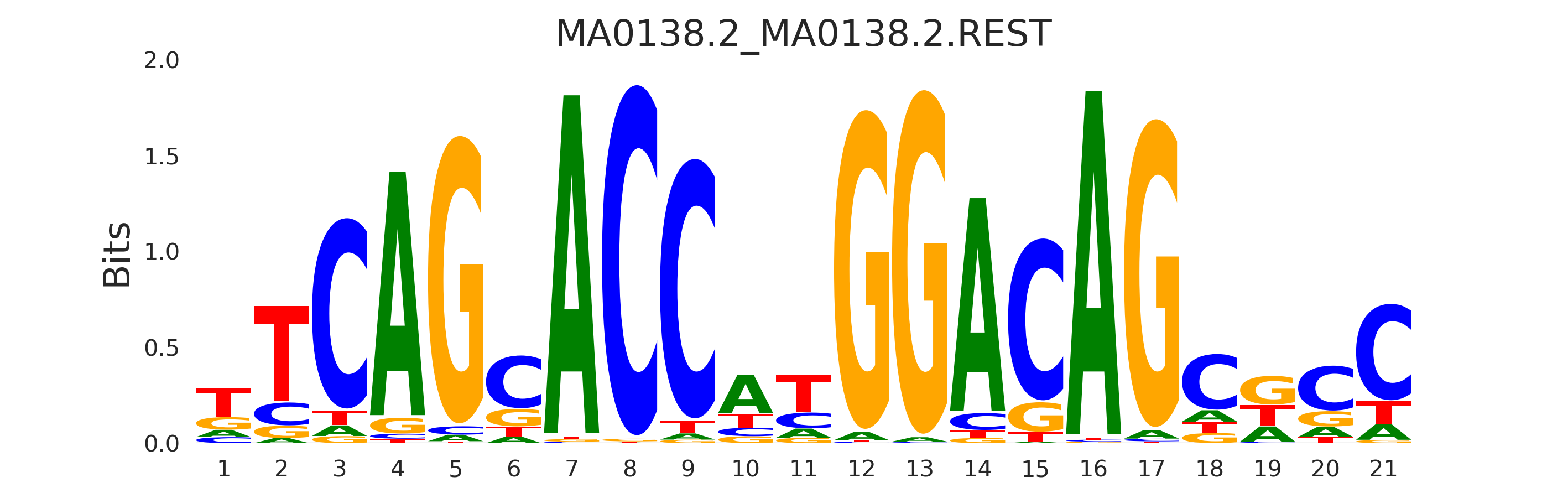
| MA0138.2_MA0138.2.REST |
MA0138.2.REST |
|
0.82 |
0.81 |
0.93 |
1.13 |
1.40 |
-1.17 |
-1.90 |
2.10 |
-2.78 |
-2.06 |
-2.78 |
-1.90 |
0.99 |
2.34 |
3.01 |
2.07 |
2 |
0.09 |
0.08 |
0.09 |
0.09 |
-0.04 |
0.01 |
-0.04 |
-0.03 |
-0.06 |
-0.05 |
-0.06 |
-0.06 |
-0.02 |
-0.03 |
-0.02 |
-0.03 |
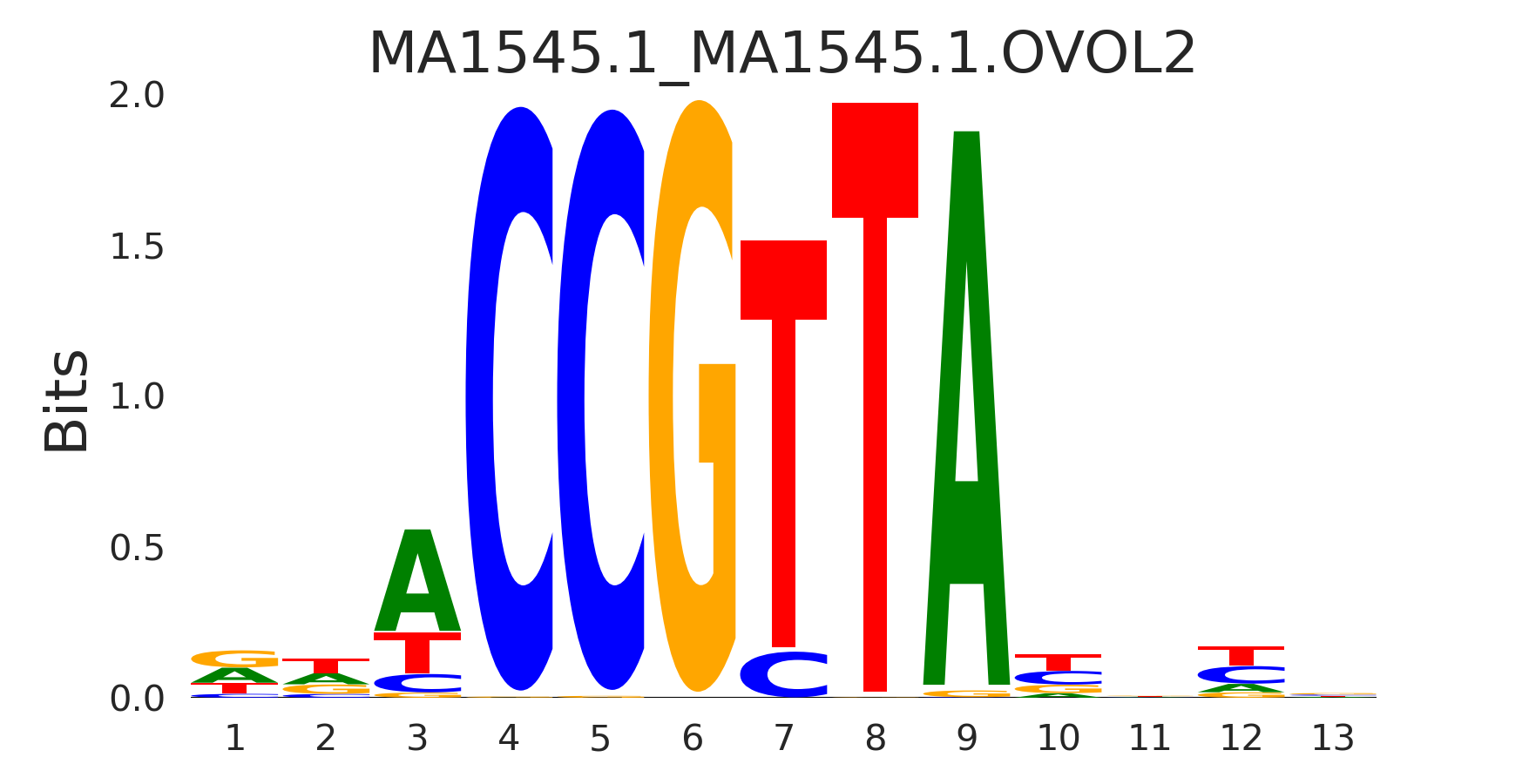
| MA1545.1_MA1545.1.OVOL2 |
MA1545.1.OVOL2,MA1544.1.OVOL1,MA0877.3.BARHL1,MA0895.1.HMBOX1,MA0776.1.MYBL1, (...) |
|
3.14 |
3.01 |
3.14 |
2.96 |
-2.86 |
3.73 |
-0.70 |
-2.37 |
0.20 |
0.52 |
0.25 |
-1.88 |
-3.34 |
-3.08 |
-3.14 |
-2.70 |
3 |
0.18 |
0.17 |
0.17 |
0.17 |
-0.14 |
0.10 |
-0.05 |
-0.12 |
0.00 |
-0.00 |
-0.01 |
-0.07 |
-0.15 |
-0.17 |
-0.22 |
-0.15 |
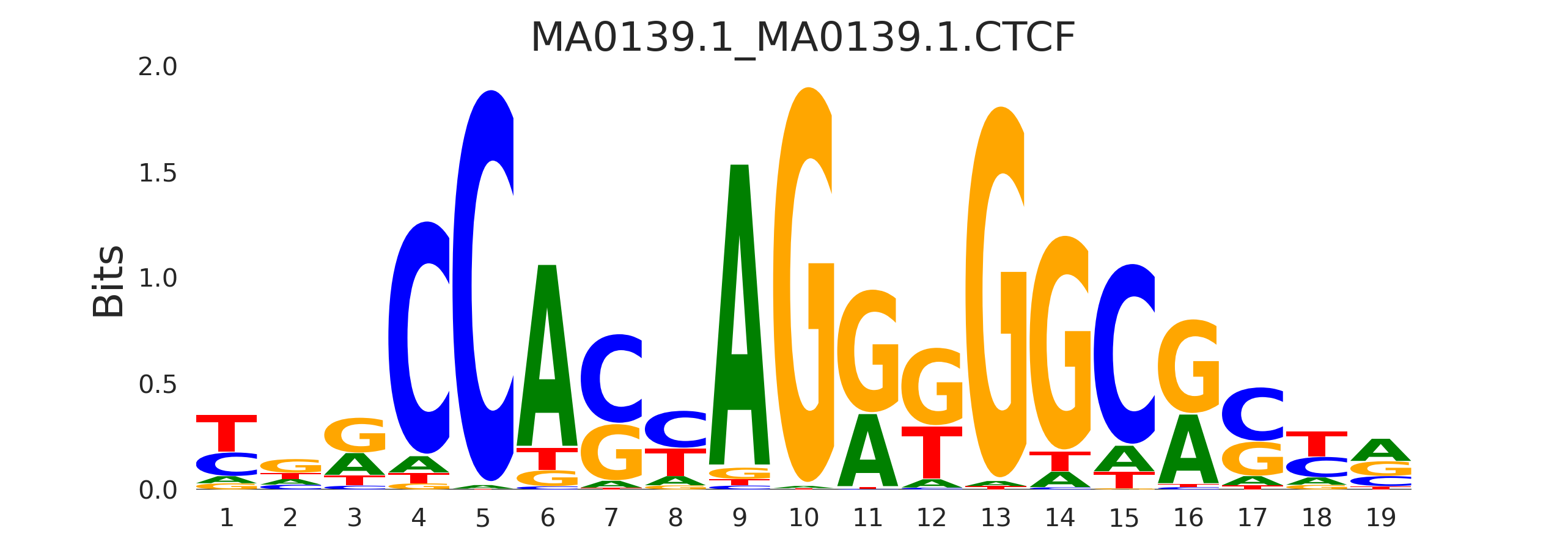
| MA0139.1_MA0139.1.CTCF |
MA0139.1.CTCF,MA1102.2.CTCFL,MA1629.1.ZIC2,MA0697.2.ZIC3,MA2025.1.CTCF, (...) |
|
2.80 |
3.08 |
2.07 |
2.70 |
-4.35 |
-2.99 |
-3.33 |
-4.35 |
3.62 |
3.01 |
2.95 |
3.50 |
-3.94 |
-3.62 |
-3.73 |
-4.35 |
3 |
0.14 |
0.14 |
0.12 |
0.13 |
-0.15 |
-0.01 |
-0.08 |
-0.20 |
0.11 |
0.06 |
0.05 |
0.04 |
-0.12 |
-0.15 |
-0.18 |
-0.16 |